Building_structural Causal_models_with_dowhy_&_econml_gcm
End-to-End approach for building Causal Models using— DoWhy + EconML + Refutation Tests + GCM API
This notebook demonstrates the main ideas and libraries used in Chapter 7 of Molak:
- DoWhy: the 4-step causal workflow
- Step 1: encode assumptions (a causal graph)
- Step 2: identify the estimand (what needs to be computed)
- Step 3: estimate the effect (linear regression, Double ML)
- Step 4: refute the estimate (robustness / stress tests)
- EconML: using Double Machine Learning (DML) as a causal estimator
- DoWhy GCM API: building a structural causal model and generating counterfactuals
Dataset choice (public + offline)
To keep this notebook runnable without external downloads, we use sklearn.datasets.load_diabetes() (ships with scikit-learn).
We then simulate a treatment and an outcome so we know the true causal effect and can sanity-check the tooling.
Why simulate treatment/outcome?
- Real public datasets rarely contain true randomized treatment.
- Chapter 7’s goal is to learn the ecosystem + workflow.
0) Setup
We try to import dowhy and econml. If they are missing, install them.
If you are in a restricted environment without pip access, run this notebook where you can pip install.
import sys
import subprocess
def pip_install(pkg):
print(f"Installing: {pkg}")
subprocess.check_call([sys.executable, "-m", "pip", "install", "-q", pkg])
# Core deps
for pkg in ["dowhy", "econml", "scikit-learn", "pandas", "numpy", "matplotlib", "networkx"]:
try:
__import__(pkg.split("==")[0])
except Exception:
pip_install(pkg)
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
Installing: dowhy
Installing: econml
Installing: scikit-learn
import warnings
from sklearn.exceptions import DataConversionWarning
warnings.simplefilter("ignore", DataConversionWarning)
1) Create a public dataset + simulate a causal story
We start with the Diabetes dataset covariates W.
Then we create:
- Treatment
X: depends on some covariates (confounding) - Outcome
Y: depends onX+ covariates
We choose a true causal effect of X → Y to be about 0.7 (like the chapter’s synthetic example).
from sklearn.datasets import load_diabetes
from sklearn.preprocessing import StandardScaler
rng = np.random.default_rng(42)
data = load_diabetes(as_frame=True)
W = data.frame.copy()
# Standardize covariates so our simulation coefficients are stable
scaler = StandardScaler()
W_scaled = pd.DataFrame(scaler.fit_transform(W), columns=W.columns)
n = len(W_scaled)
# Simulate a confounder-like latent signal using observed covariates
u = rng.normal(0, 1, size=n)
# Treatment X depends on covariates (confounding)
# (Continuous treatment to match the chapter’s DML regression example)
X = (
0.6 * W_scaled["bmi"].to_numpy() +
0.3 * W_scaled["bp"].to_numpy() +
0.2 * W_scaled["s5"].to_numpy() +
0.3 * u +
rng.normal(0, 0.5, size=n)
)
TRUE_ATE = 0.7
# Outcome Y depends on X and covariates
Y = (
TRUE_ATE * X +
0.4 * W_scaled["bmi"].to_numpy() +
0.2 * W_scaled["age"].to_numpy() -
0.3 * W_scaled["s1"].to_numpy() +
0.2 * u +
rng.normal(0, 1.0, size=n)
)
df = W_scaled.copy()
df["X"] = X
df["Y"] = Y
df.head()
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | target | X | Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.800500 | 1.065488 | 1.297088 | 0.459841 | -0.929746 | -0.732065 | -0.912451 | -0.054499 | 0.418531 | -0.370989 | -0.014719 | 1.272287 | 0.816080 |
| 1 | -0.039567 | -0.938537 | -1.082180 | -0.553505 | -0.177624 | -0.402886 | 1.564414 | -0.830301 | -1.436589 | -1.938479 | -1.001659 | -0.754342 | -0.834617 |
| 2 | 1.793307 | 1.065488 | 0.934533 | -0.119214 | -0.958674 | -0.718897 | -0.680245 | -0.054499 | 0.060156 | -0.545154 | -0.144580 | 0.590729 | 2.828625 |
| 3 | -1.872441 | -0.938537 | -0.243771 | -0.770650 | 0.256292 | 0.525397 | -0.757647 | 0.721302 | 0.476983 | -0.196823 | 0.699513 | -0.738321 | -1.308935 |
| 4 | 0.113172 | -0.938537 | -0.764944 | 0.459841 | 0.082726 | 0.327890 | 0.171178 | -0.054499 | -0.672502 | -0.980568 | -0.222496 | -0.507214 | -3.555118 |
Quick sanity check
- If we run a naive regression of Y on X without controlling for confounders, it can be biased.
- If we control for the right covariates, we should recover something close to the true effect (~0.7).
import statsmodels.api as sm
# Naive
X_naive = sm.add_constant(df[["X"]])
naive = sm.OLS(df["Y"], X_naive).fit()
print("Naive OLS coef on X:", naive.params["X"])
# Adjusted (include a bunch of covariates)
controls = [c for c in df.columns if c not in ["X", "Y"]]
X_adj = sm.add_constant(df[["X"] + controls])
adj = sm.OLS(df["Y"], X_adj).fit()
print("Adjusted OLS coef on X:", adj.params["X"])
print("True effect:", TRUE_ATE)
Naive OLS coef on X: 1.027198098583941
Adjusted OLS coef on X: 0.9013676299101012
True effect: 0.7
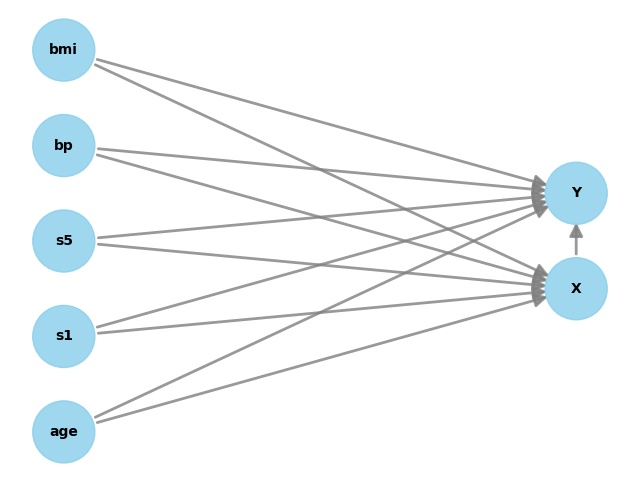
2) DoWhy Step 1 — Encode assumptions (DAG)
We now write a causal graph.
We’ll use a simple and realistic assumption:
- Several covariates W affect both X and Y (confounders)
- X affects Y
In DoWhy, you provide the graph as a string.
from dowhy import CausalModel
# We'll include a small subset of covariates to keep the graph readable.
confounders = ["bmi", "bp", "s5", "age", "s1"]
# Build a graph string (DOT format for DoWhy)
# Edges: confounders -> X and confounders -> Y, plus X -> Y
edges = []
for w in confounders:
edges.append(f"{w} -> X")
edges.append(f"{w} -> Y")
edges.append("X -> Y")
graph = "digraph {\n" + ";\n".join(edges) + ";\n}"
print(graph)
from dowhy import CausalModel
model = CausalModel(
data=df[["Y", "X"] + confounders],
treatment="X",
outcome="Y",
graph=graph
)
model.view_model()
digraph {
bmi -> X;
bmi -> Y;
bp -> X;
bp -> Y;
s5 -> X;
s5 -> Y;
age -> X;
age -> Y;
s1 -> X;
s1 -> Y;
X -> Y;
}
3) DoWhy Step 2 — Identify the estimand
Now DoWhy uses the graph to answer:
“What expression can I compute from data to get the causal effect of X on Y?”
Usually, you’ll see it choose a backdoor adjustment set (control variables).
estimand = model.identify_effect()
print(estimand)
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
────(E[Y|age,s1,s5,bp,bmi])
d[X]
Estimand assumption 1, Unconfoundedness: If U→{X} and U→Y then P(Y|X,age,s1,s5,bp,bmi,U) = P(Y|X,age,s1,s5,bp,bmi)
### Estimand : 2
Estimand name: iv
No such variable(s) found!
### Estimand : 3
Estimand name: frontdoor
No such variable(s) found!
### Estimand : 4
Estimand name: general_adjustment
Estimand expression:
d
────(E[Y|age,s1,s5,bp,bmi])
d[X]
Estimand assumption 1, Unconfoundedness: If U→{X} and U→Y then P(Y|X,age,s1,s5,bp,bmi,U) = P(Y|X,age,s1,s5,bp,bmi)
4) DoWhy Step 3 — Estimate the effect
We estimate the causal effect using two estimators (like the chapter):
- Linear regression (simple baseline)
- Double ML (DML) via EconML (more flexible, ML-based)
We then compare results to the known true effect (~0.7).
# 4.1 Linear regression estimator inside DoWhy
estimate_lr = model.estimate_effect(
identified_estimand=estimand,
method_name="backdoor.linear_regression"
)
print("DoWhy Linear Regression ATE:", estimate_lr.value)
print("True effect:", TRUE_ATE)
DoWhy Linear Regression ATE: 0.8933354974909335
True effect: 0.7
# 4.2 Double ML estimator via EconML
# We'll use gradient boosting to model the nuisance functions
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.linear_model import LassoCV
estimate_dml = model.estimate_effect(
identified_estimand=estimand,
method_name="backdoor.econml.dml.DML",
method_params={
"init_params": {
"model_y": GradientBoostingRegressor(random_state=42),
"model_t": GradientBoostingRegressor(random_state=42),
"model_final": LassoCV(fit_intercept=False, random_state=42),
},
"fit_params": {}
}
)
print("DoWhy + EconML DML ATE:", estimate_dml.value)
print("True effect:", TRUE_ATE)
DoWhy + EconML DML ATE: 0.9479973316124858
True effect: 0.7
Interpretation
- If both estimates are close to ~0.7, our identification + estimation is behaving properly.
- In real projects, you don’t know the true effect — so you rely more on:
- refutation tests
- sensitivity checks
- domain validation
5) DoWhy Step 4 — Refute the estimate (robustness tests)
Refutation tests try to break your causal result.
We’ll run the same style of tests used in the chapter:
- Data subset refuter (invariant transformation)
- Random common cause (invariant transformation)
- Placebo treatment refuter (nullifying transformation)
Key idea:
- Invariant transformations should not change the estimate much.
- Nullifying transformations should make the effect go to ~0.
# We'll refute the DML estimate (you can also refute the linear one)
print("Original DML estimate:", estimate_dml.value)
Original DML estimate: 0.9479973316124858
# 5.1 Data subset refuter
refute_subset = model.refute_estimate(
estimand=estimand,
estimate=estimate_dml,
method_name="data_subset_refuter",
subset_fraction=0.4,
random_seed=42
)
print(refute_subset)
Refute: Use a subset of data
Estimated effect:0.9479973316124858
New effect:0.8400437564259516
p value:0.48
# 5.2 Random common cause
refute_random_cc = model.refute_estimate(
estimand=estimand,
estimate=estimate_dml,
method_name="random_common_cause",
random_seed=42
)
print(refute_random_cc)
Refute: Add a random common cause
Estimated effect:0.9479973316124858
New effect:0.8424992466817046
p value:0.08
# 5.3 Placebo treatment refuter
refute_placebo = model.refute_estimate(
estimand=estimand,
estimate=estimate_dml,
method_name="placebo_treatment_refuter",
placebo_type="permute",
random_seed=42
)
print(refute_placebo)
Refute: Use a Placebo Treatment
Estimated effect:0.9479973316124858
New effect:0.003774568832559745
p value:0.98
How to read refutation outputs
Typically you see:
- Estimated effect (original)
- New effect (after refutation)
- p-value for the difference
What you want:
- subset/common-cause: new effect ≈ old effect (difference not significant)
- placebo: new effect ≈ 0
6) GCM API (extra) — structural causal modeling + counterfactuals
DoWhy’s GCM API lets you go beyond ATE and ask:
- “How strong are the incoming causal arrows into Y?”
- “What would Y have been if we set X to another value (counterfactual)?”
This requires specifying a causal mechanism for each node.
We’ll build a small graph with the same variables used above.
import networkx as nx
from dowhy import gcm
from sklearn.linear_model import LinearRegression
# Build the same causal structure using NetworkX
G = nx.DiGraph()
for w in confounders:
G.add_edge(w, "X")
G.add_edge(w, "Y")
G.add_edge("X", "Y")
# Plot (optional)
pos = nx.spring_layout(G, seed=42)
nx.draw(G, pos, with_labels=True)
plt.show()
from dowhy import gcm
from sklearn.linear_model import LinearRegression
from dowhy.gcm.ml import SklearnRegressionModel
# Create an invertible SCM (good for counterfactuals)
causal_model = gcm.InvertibleStructuralCausalModel(G)
# Root nodes as empirical distributions
for w in confounders:
causal_model.set_causal_mechanism(w, gcm.EmpiricalDistribution())
# X and Y as additive noise models with wrapped sklearn regressors
causal_model.set_causal_mechanism("X", gcm.AdditiveNoiseModel(SklearnRegressionModel(LinearRegression())))
causal_model.set_causal_mechanism("Y", gcm.AdditiveNoiseModel(SklearnRegressionModel(LinearRegression())))
# Fit mechanisms to data
gcm.fit(causal_model, df[["X", "Y"] + confounders])
print("GCM fitted.")
Fitting causal mechanism of node s1: 100%|██████████████████████████████████████████████| 7/7 [00:00<00:00, 282.99it/s]
GCM fitted.
6.1 Arrow strength into Y
This gives a measure of how much each parent contributes to explaining Y. This is not exactly the same as ATE. It’s more like: “How important is this causal parent in the model for Y?”
strengths = gcm.arrow_strength(causal_model, "Y")
strengths
{('X', 'Y'): np.float64(0.7045566616574985),
('age', 'Y'): np.float64(0.04788431176770035),
('bmi', 'Y'): np.float64(0.15649327627968032),
('bp', 'Y'): np.float64(0.01957459130750313),
('s1', 'Y'): np.float64(0.08355406661605072),
('s5', 'Y'): np.float64(0.009460206774970321)}
6.2 Counterfactual example
Counterfactual question:
For a specific observation, what would Y be if we forced X to some value?
We pick one row and set X to a new value.
row = df[["X", "Y"] + confounders].iloc[[0]].copy()
row
| X | Y | bmi | bp | s5 | age | s1 | |
|---|---|---|---|---|---|---|---|
| 0 | 1.272287 | 0.81608 | 1.297088 | 0.459841 | 0.418531 | 0.8005 | -0.929746 |
# Force X to a new value (intervention)
new_x = float(row["X"].iloc[0] - 1.0) # just as an example
cf = gcm.counterfactual_samples(
causal_model,
interventions={"X": lambda _: new_x},
observed_data=row
)
print("Observed:")
print(row[["X", "Y"] + confounders])
print("Counterfactual (X forced to new value):")
print(cf[["X", "Y"] + confounders])
Observed:
X Y bmi bp s5 age s1
0 1.272287 0.81608 1.297088 0.459841 0.418531 0.8005 -0.929746
Counterfactual (X forced to new value):
X Y bmi bp s5 age s1
0 0.272287 -0.077256 1.297088 0.459841 0.418531 0.8005 -0.929746
7) What you should take away (Chapter 7 in one page)
- DoWhy gives you a clean 4-step causal workflow: 1) graph → 2) estimand → 3) estimate → 4) refute
- EconML lets you plug in modern estimators like DML when you need ML flexibility.
- Refutation tests are your “validation set” for causal claims (they try to break the result).
- GCM lets you move into structural models and ask counterfactual questions.
Practical advice
In real marketing/personalization work, a solid minimum deliverable is:
- DAG + explanation of assumptions
- at least one backdoor estimator (linear or DML)
- refutation suite (subset, placebo, random common cause)
- sensitivity check (if possible)